Software
When collecting air quality measurements, one of the main goals is often to identify the sources of those compounds. This is done through source apportionment, a general term for any method that organizes measurements into distinct source profiles. The most common approach is Positive Matrix Factorization (PMF), a matrix factorization technique that decomposes the observed data into source profiles or fingerprints and their relative contributions. Each fingerprint corresponds to a potential emission source.
However, assigning a real-world source to each fingerprint can be tedious and subjective. The go-to reference for known source profiles is the EPA's SPECIATE database, which contains over 6,000 profiles and 1,500 analytes. Finding which known source best matches a fingerprint requires manually comparing across this massive database which is a slow and inconsistent process.
To automate and standardize this step, I created pmf2speciate, an open-source Python package that identifies PMF source profiles using a multi-tiered random forest classifier trained on the EPA SPECIATE database. The tool speeds up source identification, reduces subjectivity, and provides a reproducible, data-driven workflow.
Data Preparation
The EPA SPECIATE dataset is not only large but it is also incredibly messy. Each profile quantifies the contribution of an analyte to a fingerprint as a percent weight (out of 100), but these data are entered manually, leading to typos, inconsistent capitalization, punctuation, and naming. For example, the same source might appear as "Biomass Burning: Wildfire," "Wildfire," or "Burning: Wildfire" in different samples. To clean and group these entries, I used a large language model (LLM) to cluster similar names into unified source categories. Say what you will about LLMs, but this was one case where they saved me days of manual cleanup.
Once profiles were grouped, I split the data into training and test sets. Because many sources had limited or uneven sample counts, I augmented the data by generating synthetic samples using each profile's reported uncertainties, or when uncertainties were unavailable, by adding Gaussian noise. This ensured the model had enough variability to generalize while maintaining realistic distributions.
Model Architecture
The classification follows a two-tier hierarchy:
- Tier 1: Classify the fingerprint by generation mechanism (e.g., combustion, volatilization, microbial, etc.).
- Tier 2: Within that generation mechanism, classify the fingerprint by specific source.
I trained a random forest classifier for each level using sklearn. Tier 1 contains one model that classifies generation mechanisms; Tier 2 contains separate models trained on the sources within each mechanism. Currently, there are three generation mechanisms, giving four total models.
Each model outputs class probabilities using sklearn's predict_proba, allowing pmf2speciate to assign a confidence score to both classification levels.
Model Training and Performance
Performance varies by source class. Sources with many well-defined profiles (like gasoline emissions or biomass burning) achieve higher accuracy, while sparse categories relying on synthetic-generated samples perform less consistently.
The overall Tier 1 model achieves strong separation between generation mechanisms, with all F1 scores above 0.95. Within each mechanism, Tier 2 models achieve macro-averaged accuracies ranging from roughly 0.6 to 0.8.
Each model's confusion matrix highlights this variability. Some classes show near-perfect normalized accuracy (1.0), which could indicate slight overfitting, especially for sources with few training samples or low chemical variability. Others perform worse, likely due to sparse data or overlapping chemical fingerprints between similar sources.
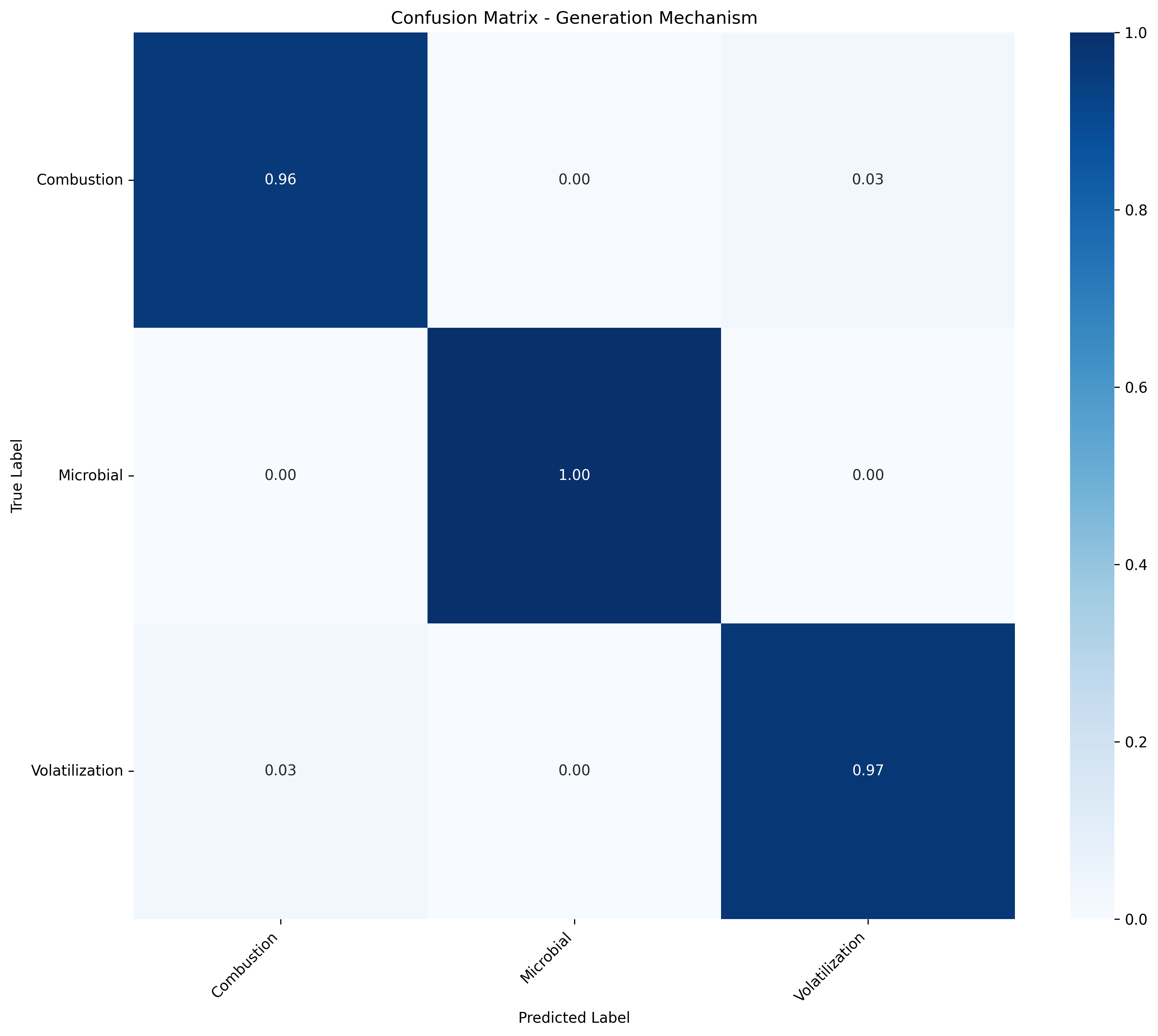
Figure 1: Tier 1 confusion matrix - Generation mechanisms
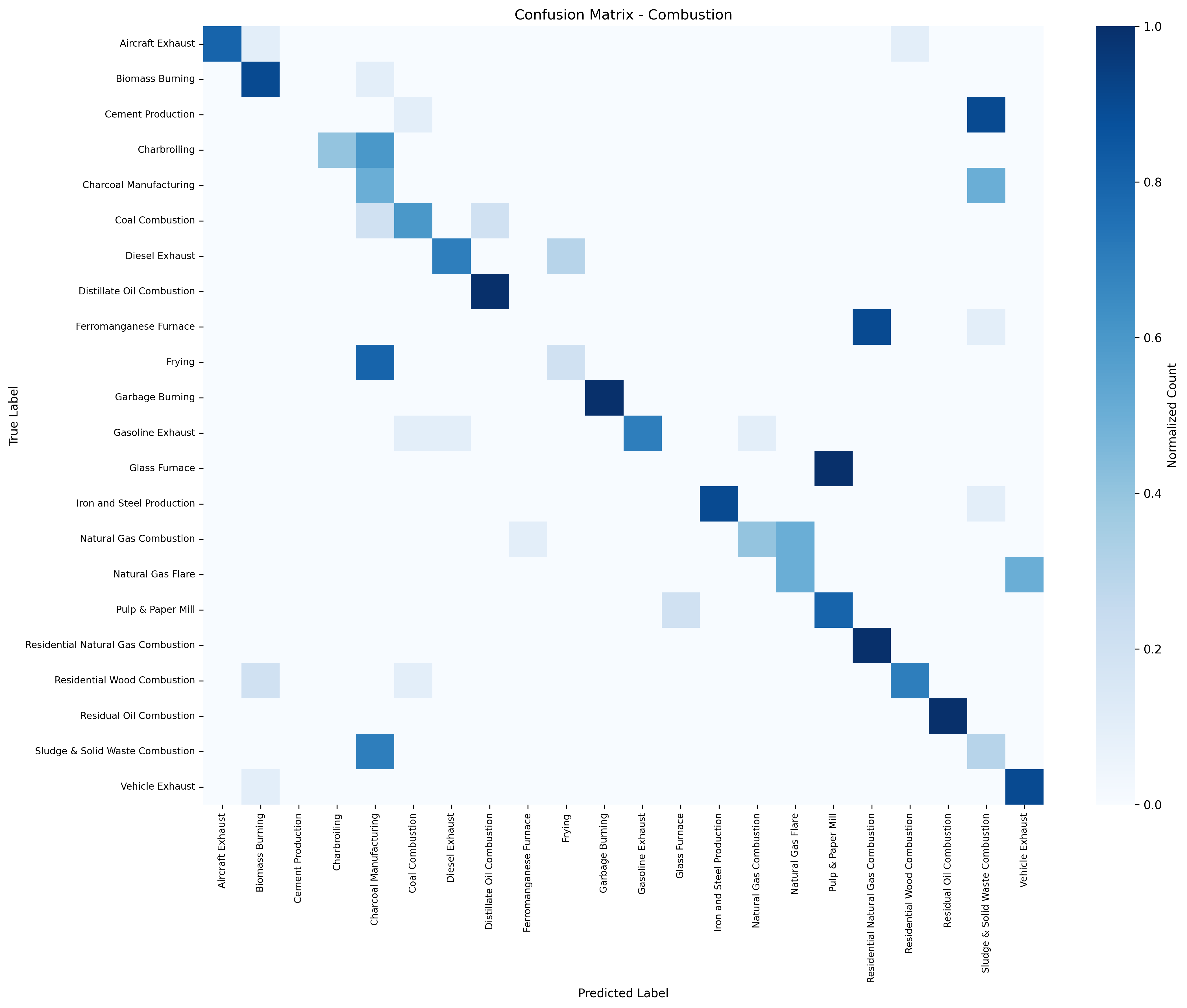
Figure 2: Tier 2 confusion matrix - Combustion sources
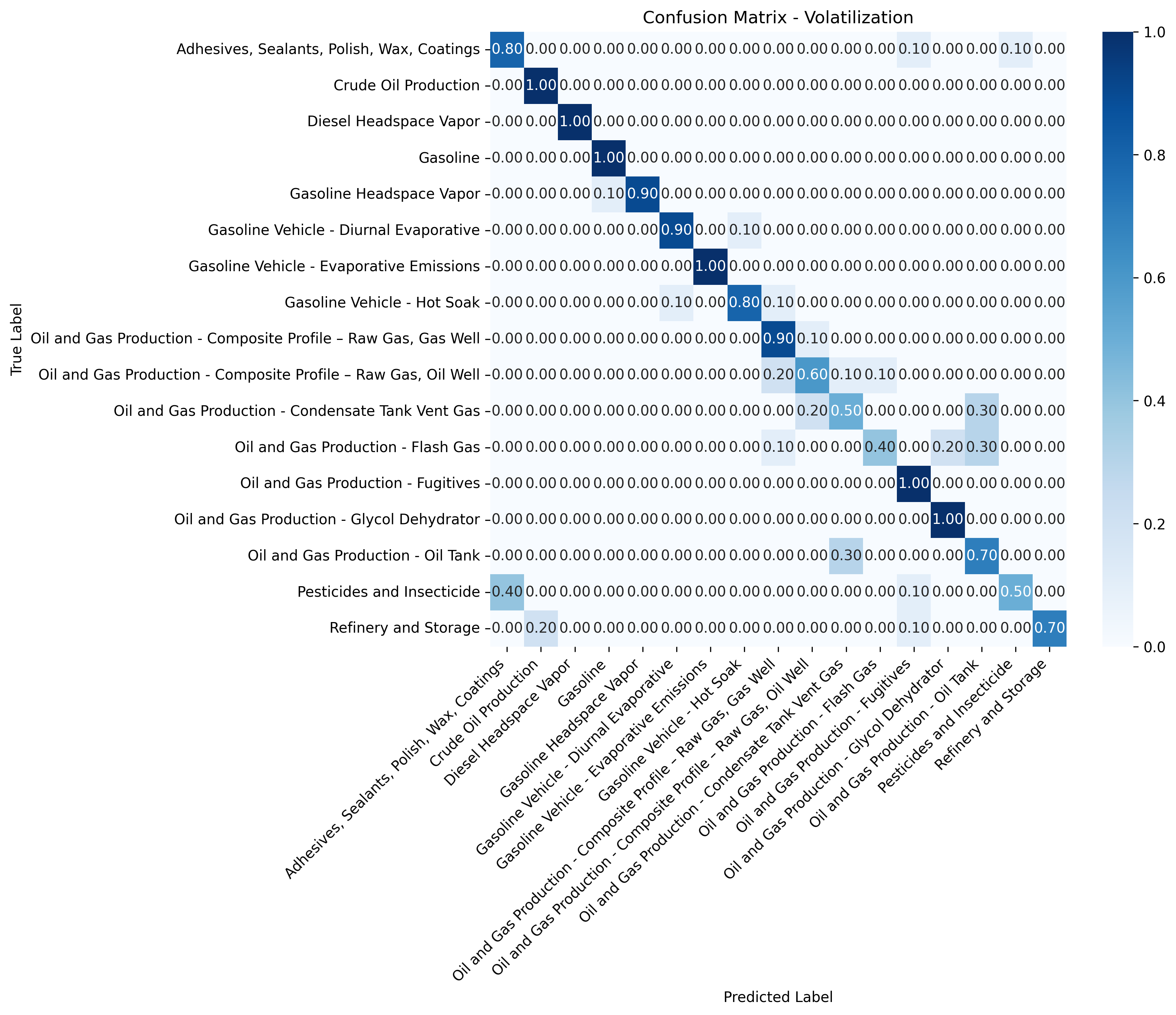
Figure 3: Tier 2 confusion matrix - Volatilization sources
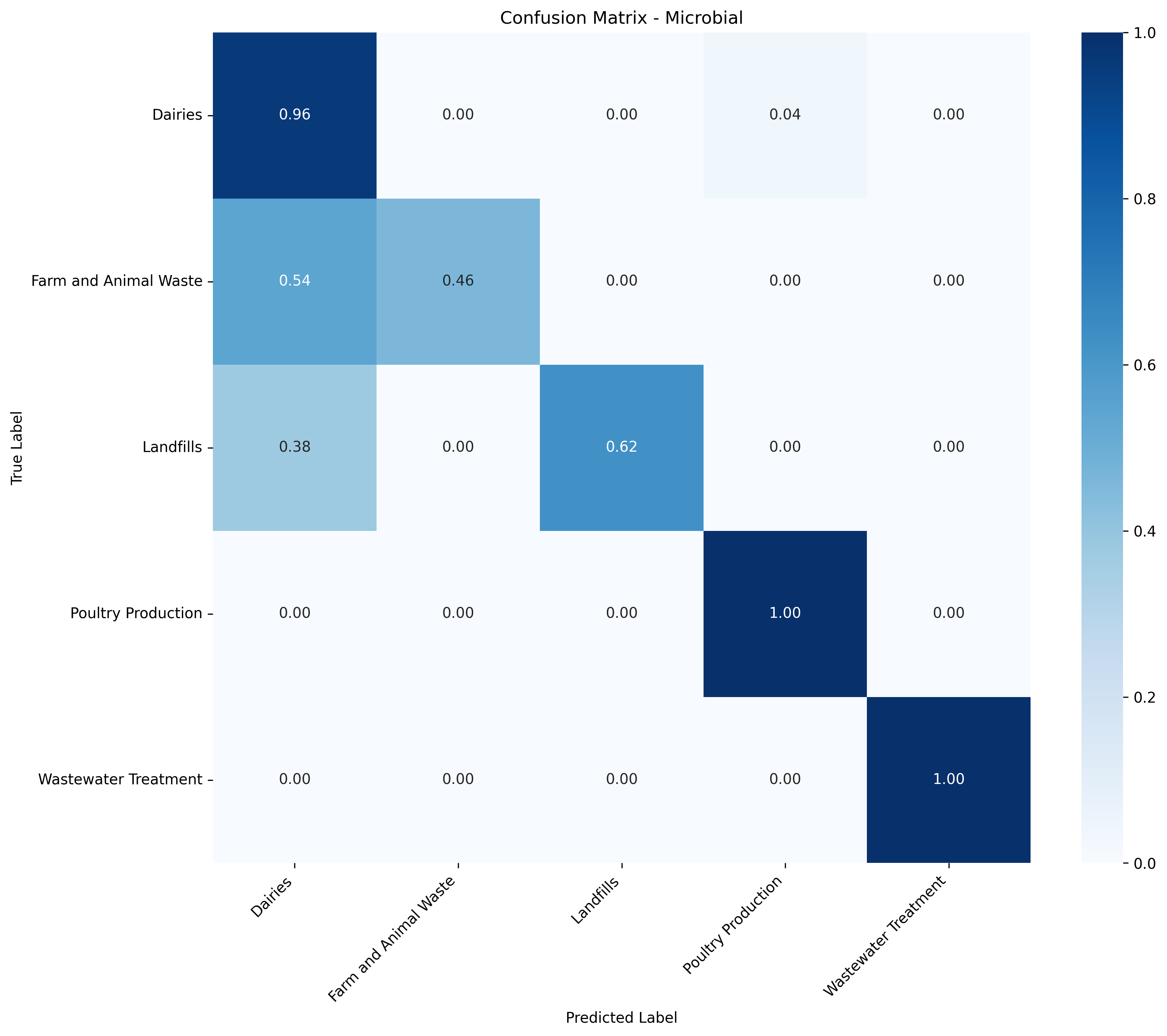
Figure 4: Tier 2 confusion matrix - Microbial sources
For now, I prioritize building a working, end-to-end system that automates source identification and returns interpretable confidence scores. The next step will be refining underperforming classes.
Package Usage
The package is designed to be easy to use. Identifying a fingerprint takes just a few lines of code:
from pmf2speciate import SourceClassifier
factor_profile = {
"71-43-2": 15.2,
"108-88-3": 8.7,
"100-41-4": 5.1,
"1330-20-7": 12.3,
# ... more species
}
classifier = SourceClassifier()
result = classifier.identify_source(factor_profile)
print("Classification Result:")
print(f"Generation Mechanism: {result['generation_mechanism']} (confidence: {result['generation_confidence']:.3f})")
if result["specific_source"]:
print(f"Specific Source: {result['specific_source']} (confidence: {result['source_confidence']:.3f})")
print(f"Overall Confidence: {result['overall_confidence']:.3f}")Fingerprints can also be compared to the average source profile:
from pmf2speciate import plot_factor
plot_factor(factor_profile, result["generation_mechanism"], result["specific_source"])This lets users visually verify whether they agree with the classification, preserving expert judgment while providing automated guidance.
Additional utilities include viewing model info:
from pmf2speciate import SourceClassifier
classifier = SourceClassifier()
print(classifier.get_model_info())and visualizing (average) source profiles within each generation mechanism:
from pmf2speciate import plot_profiles
plot_profiles("Combustion")
plot_profiles("Microbial")
plot_profiles("Volatilization")Code Availability
The pmf2speciate package is available on GitHub and can be installed via pip.